Mechanokinases
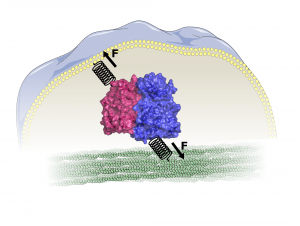
Kinases are abundantly present in all cells and involved in the regulation of many important processes, such as cell survival, growth, development and motility. They transmit biochemical signals by phosphorylating their target proteins. In the MBM group we aim to understand the force mediated activation mechanism of mechanokinases at atomistic detail using force-probe MD simulations.
Cells experience forces from their environment, such as shearing or compression. They need to sense and integrate these inputs and react accordingly for example to resist deformation. This is often achieved by remodeling of the cytoskeleton, which provides shape and support to cells. How do cells convert such mechanical into biochemical cues, which can be communicated within the cells? This is where kinases come into play. Some not only transmit signals, but also sense forces and become activated by them. These mechanosensing kinases are found at adhesions sites where cells bind to neighboring cells or the extracellular matrix. They have a connection – either direct or through other proteins – to the cellular membrane and to the cytoskeleton and they are inhibited by another protein or a domain of the same protein. Forces acting on cells can release these (auto-)inhibitory interactions.
Examples include focal adhesion kinase (FAK) or Src kinase. We simulated forces acting on FAK’s binding sites to the cellular membrane and cytoskeleton link, which are able to open the interface between its autoinhibitory FERM- and its kinase domain and make the kinase active site accessible.
For Src, we are investigating whether force acting through its fatty acid – membrane linkage and its binding partner p130Cas impairs autoinhibition by its SH domains.
A special kinase that we are interested in is integrin-linked kinase (ILK), which is considered as a pseudokinase. Thus it does not perform biochemical catalysis but still binds ATP. Using force-probe MD and cellular experiments we investigate the function of ATP on ILK mechanotransduction, binding to its partner protein parvin and cellular force generation.
For details on FAK see:
Acebrón I, Righetto RD, Schoenherr C, de Buhr S, Redondo P, Culley J, Rodríguez CF, Daday C, Biyani N, Llorca O, Byron A, Chami M, Gräter F, Boskovic J, Frame MC, Stahlberg H, Lietha D (2020). Structural basis of Focal Adhesion Kinase activation on lipid membranes, EMBO J 39:e104743 (https://doi.org/10.15252/embj.2020104743)
Bauer MS, Baumann F, Daday C, Redondo P, Durner E, Jobst MA, Milles LF, Mercadante D, Pippig DA, Gaub HE, Gräter F, Lietha D (2019). Structural and mechanistic insights into mechanoactivation of focal adhesion kinase, Proc Natl Acad Sci USA 116(14):6766-6774 (https://doi.org/10.1073/pnas.1820567116)
Zhou J, Aponte-Santamaría C, Sturm S, Bullerjahn JT, Bronowska A, Gräter F (2015). Mechanism of Focal Adhesion Kinase Mechanosensing, PLoS Comput Biol, 11(11):e1004593 (https://doi.org/10.1371/journal.pcbi.1004593)