Protein structures in systems biology
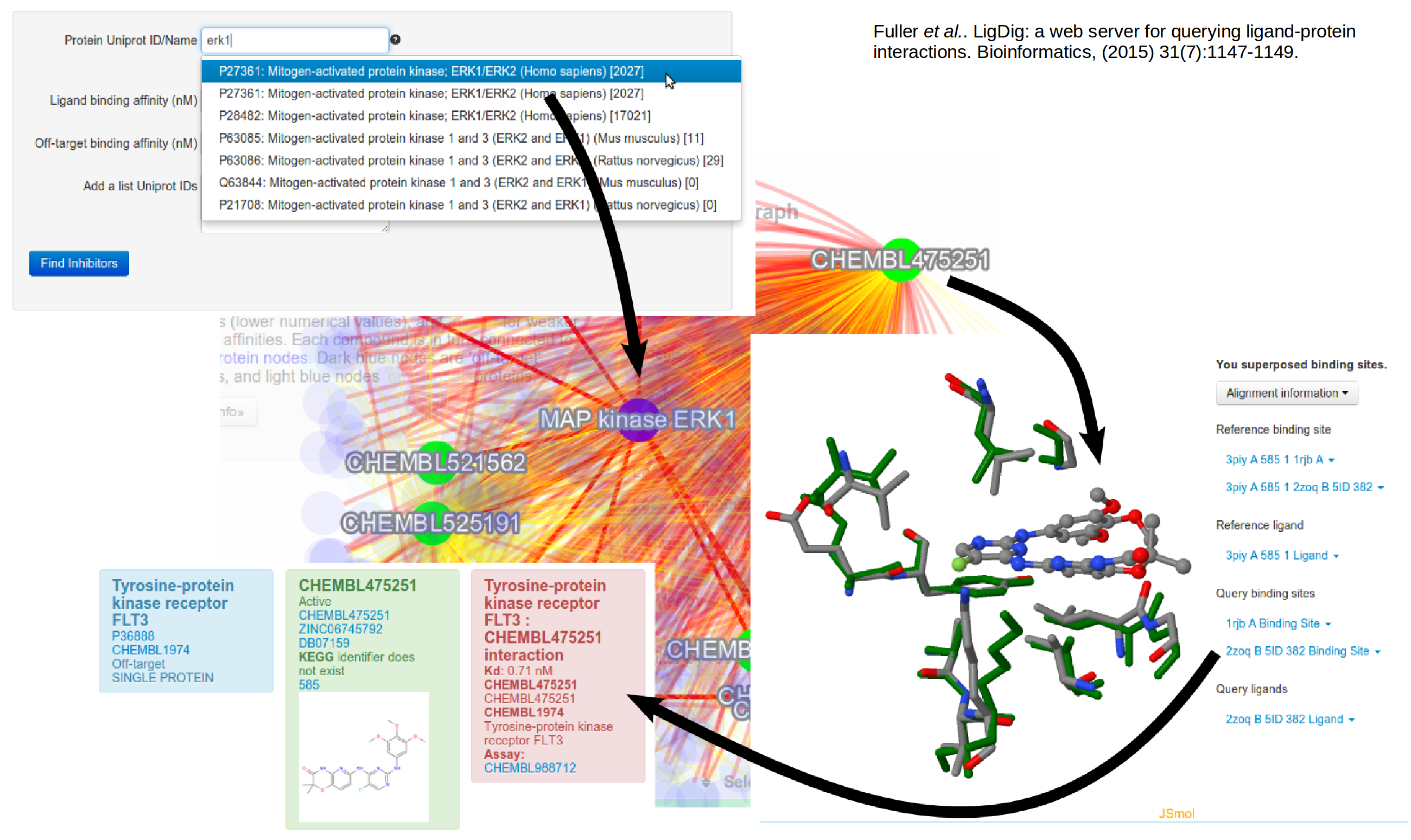
We are working on developing approaches to bridge between protein structures and biochemical networks, from the molecular to the cellular level.
In the Human Brain Project, we are contributing to the development of multi-scale molecular simulation techniques for the brain simulation platform, EBRAINS. An example of the application of protein structure-based molecular dynamics simulations, bioinformatics analysis and mathematical modeling of signalling networks is given by a study of how regulation of the enzyme adenylyl cyclase 5 in striatal neurons confers the ability to detect coincident neuromodulatory signals, which is important for reward learning (Bruce et al., 2019).
In the Sysmolab (Comparative systems biology of lactic acid bacteria) projects, we explored how differences in the allosteric regulation of key metabolic enzymes can be related to differences in the environment in which the bacteria live (Feldman-Salit, 2013; Veith, 2013). These studies made use of our PIPSA (Protein Interaction Property Similarity Analysis) webserver and software, as well as SYCAMORE (SYstems biology’s Computational Analysis and MOdeling Research Environment) .