EnzymeML: Surfing the research data wave

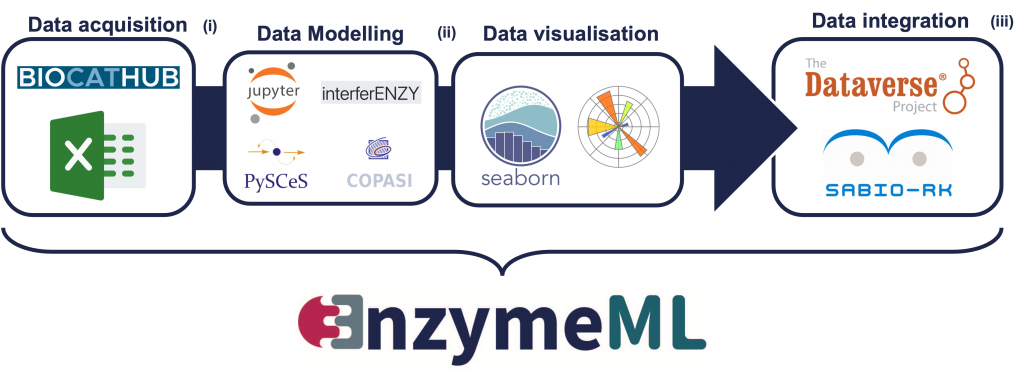
In catalytic sciences we face a rapidly increasing volume and complexity of research data, which are a challenge for analysis and reuse. A team of researchers and database experts has introduced EnzymeML as a data exchange format, in an article in “Nature Methods”. EnzymeML serves as a format to comprehensively report the results of an enzymatic experiment and stores the data in a structured way and makes it traceable and reusable. HITS researchers contribute to this project with the SABIO-RK database that stores kinetic data directly after they have been modeled.
While more and more data is generated by an increasing number of researchers and increasing research expenditure worldwide, this data is hardly manageable by our scholarly practice of communicating scientific results. The lack of standards, incomplete metadata, and missing original data make it nearly impossible to reproduce published results. This also applies to studies on the catalytic activity, selectivity and stability of enzymes and enzymatic networks, a field of research that is equally important for industrial biotechnology and biomedicine. Moreover, data describing enzymatic experiments is particularly complex, because an enzymatic reaction depends on many factors, such as the protein sequence of the Enzyme, or the pH value.
The new, standardized data exchange format “EnzymeML”, presented by 23 authors from 14 different research institutions in the scientific journal “Nature Methods” gives hope in this respect. EnzymeML can completely record the results of an enzymatic experiment, from the reaction conditions to the measured data, as well as the kinetic model used to analyze experimental data and the estimated kinetic parameters. The
format thus provides a seamless communication channel between experimental platforms, electronic lab notebooks, enzyme kinetics modeling tools, publication platforms, and enzymatic reaction databases.
The project is coordinated by Jürgen Pleiss, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, who is also the corresponding author. HITS researchers Ulrike Wittig and Andreas Weidemann are in charge of the data integration in EnzymeML. They use SABIO-RK, a reaction kinetics database developed at HITS, where the kinetic data will be stored directly after they have been modeled.
More details in this press release, issued by the University of Stuttgart: https://www.uni-stuttgart.de/en/university/news/all/Surfing-the-research-data-wave/
Original publication:
Simone Lauterbach, et al.: EnzymeML: “Seamless data flow and modeling of enzymatic data”, Nature Methods 2023, DOI 10.1038/s41592-022-01763-1
Expert contact:
University of Stuttgart:
Prof. Dr. Jürgen Pleiss, University of Stuttgart
Tel.: +49 711 685 63191
email:juergen.pleiss@itb.uni-stuttgart.de
HITS:
Dr. Ulrike Wittig
Press contact:
University of Stuttgart:
Andrea Mayer-Grenu, University
Communications,
Tel.: +49 (0)711/685 82176
Email: andrea.mayergrenu@hkom.uni-stuttgart.de
HITS:
Dr. Peter Saueressig
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.