Simulation of transmembrane proteins in lipid environments: not always an easy task
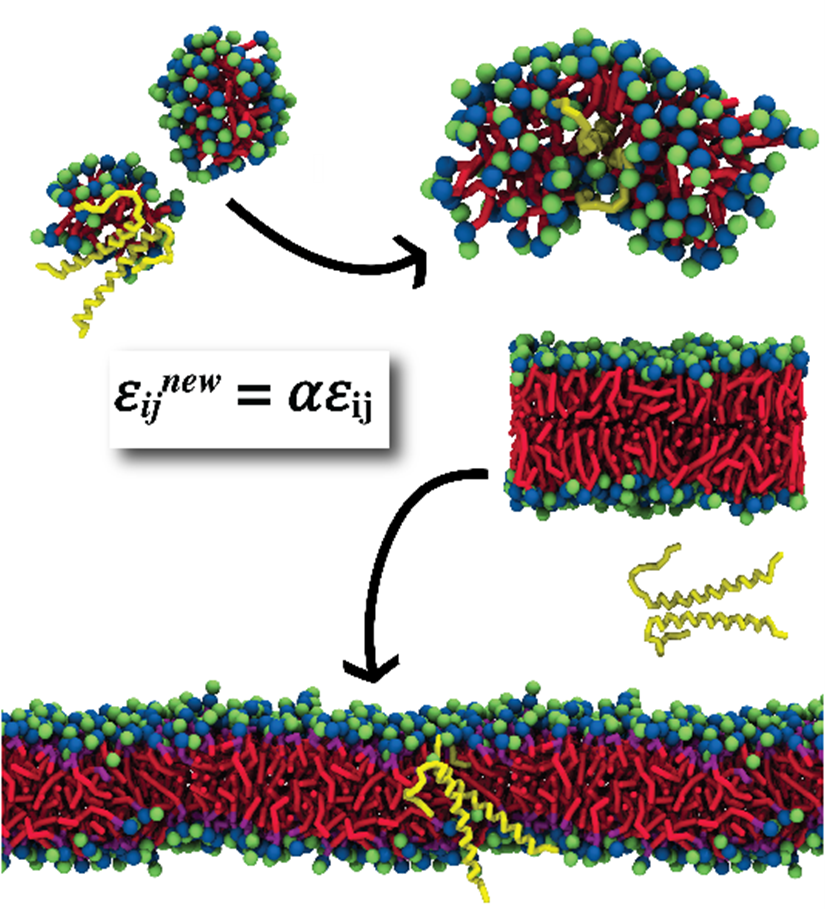
Researchers in the Molecular and Cellular Modeling group (MCM) at HITS have recently released the first parameters of a micelle-forming lipid for the widely used coarse grained force field Martini 3. The lipid they have parameterized is dodecylphosphocholine (DPC), which is of interest for the scientific community because it is often used in experiments on transmembrane proteins.
Simulations of DPC with these parameters showed micelle properties very similar to those obtained in experiments. However, these simulations showed something unexpected as well: transmembrane protein dimers were not encapsulated in lipid micelles, but rather interacted with the water in the solvent phase. This behavior is not expected for such hydrophobic polymers, so the researchers started to look at this problem more in detail.
A more fundamental issue
First, they simulated different proteins to assess the generality of this behavior. They chose the model helical dimer glycophorin A and the transmembrane region of the TrkA neurotrophin receptor, which have a similar structure but different amino acid sequences. As neither of these proteins were encapsulated in DPC micelles, they hypothesized that there might be a problem with the new DPC parameters. To test this possibility, the researchers ran bilayer self-assembly simulations with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, a phospholipid that had previously been parameterized and whose behavior is well-understood. Surprisingly, the protein failed to be incorporated in the lipid bilayer, pointing to a more fundamental issue.
With the aim of finding a simple and effective solution to this problem, the researchers tried to increase the strength of the protein-lipid interactions to stabilize the proteins in the bilayer or micelle. However, these attempts were unsuccessful, so they used the opposite approach and scaled down the protein-water interactions by 10%. This resolved the issue, enabling transmembrane proteins to be encapsulated into micelles and bilayers during self-assembly. The researchers recommend using this method for simulations of transmembrane proteins in micelles or for self-assembling bilayers with proteins using Martini 3.
The findings have been published in the Journal of Chemical Theory and Computation.
Scaling Protein–Water Interactions in the Martini 3 Coarse-Grained Force Field to Simulate Transmembrane Helix Dimers in Different Lipid Environments. Ainara Claveras Cabezudo, Christina Athanasiou, Alexandros Tsengenes, and Rebecca C. Wade. Journal of Chemical Theory and Computation 2023 19 (7), 2109-2119 DOI: 10.1021/acs.jctc.2c00950
The research was supported by the European Union’s Horizon 2020 research and innovation programme “Euroneurotrophin” under the Marie Skłodowska-Curie grant agreement No 765704 and the Klaus Tschira Foundation and made use of computing resources provided by the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through Grant INST 35/1134-1 FUGG.
Scientific contact:
Prof. Dr. Rebecca Wade
Molecular and Cellular Modeling group (MCM)
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.